Recently I have been starting to use dplyr for handling my data in R. It makes everything a lot smoother! My previous workflow – running an SQL query, storing the results as CSV, loading it in RStudio – is now history. With dplyr you can directly query data from many different databases in a very convenient way. Unfortunately Microsoft SQL Server is not directly supported but by using the package RSQLServer it can be done like with any other database. In this blog post I’ll explain how I installed everything on my Windows 7 machine to access MSSQL Server with R, since it was not as straight forward as one might think.
Contents
All you need … theoretically
The package RSQLServer is not available on CRAN anymore but it can be installed from the github repo imanuelcostigan/RSQLServer.
# install.packages('devtools')
devtools::install_github('imanuelcostigan/RSQLServer')
If this works you’re lucky and already have all the necessary things installed. If not, follow the steps below 🙂
Errors I encountered
On my path from finding out about RSQLServer to actually using it I had to fix a few things. Searching on the internet and finding most of the answers on stackoverflow I was able to piece together everything I needed. To save everyone else who tries this some time I collected everything I found out.
Fixing the installation
The first error occured when I tried to install the package as shown above.
Downloading GitHub repo imanuelcostigan/RSQLServer@master
from URL https://api.github.com/repos/imanuelcostigan/RSQLServer/zipball/master
Installing RSQLServer
Downloading GitHub repo hadley/dplyr@63d4a9f5
from URL https://api.github.com/repos/hadley/dplyr/zipball/63d4a9f5
Error: running command ‘”C:/PROGRA~1/R/R-33~1.2/bin/x64/R” –no-site-file –no-environ –no-save –no-restore –quiet CMD config CC’ had status 1
To resolve this:
- Find out which R version you have, if you don’t know, type
R.versioninside an R session - Download and install the correct version of Rtools
- Although I had R version 3.3 and downloaded the correct R version for it, it tried to install it in a directory called “34”. To make sure it will later find it, I changed this to “33” to match my R version. I am not sure whether it made a difference.
- Make sure to check the option where it asks if you want to add it to the path. This makes a difference.
- Make sure you have the correct Java version and/or architecture installed. I didn’t have Java installed for x86 which also caused the installation to fail.
- Restart your computer or at least log out and in again.
After that I was able to install the package using the following command.
devtools::install_github('imanuelcostigan/RSQLServer')
Getting integrated (NTLM) authentication to work
Download jtds-drivers and follow these instructions.
Finally accessing MSSQL Server with R
I was working with two different databases. One was remote and with that it worked immediately. The other one was a local installation on my computer.
For the local one I got an error message. Note: I don’t think it had anything to do with the databases being remote/local, just with the settings I used for installing my database!
Error in rJava::.jcall(driver@jdrv, “Ljava/sql/Connection;”, “connect”, :
java.sql.SQLException: Network error IOException: Connection refused: connect
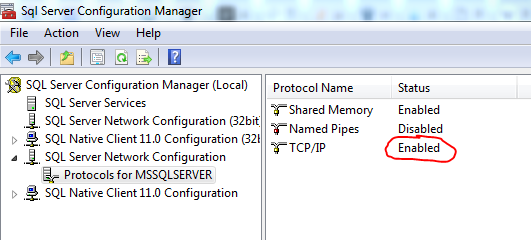
To resolve this, you need to open the SQL Server configuration manager on the machine where SQL Server is running. I have the SQL Server configuration manager and the SQL Server configuration manager (2016) installed. For some settings it doesn’t matter which one you use but for this one I had to use the older one otherwise my database (SQL Server 2012) wouldn’t show up.
Make sure to enable TCP/IP. Restart you SQL Server instance. Now it should be working!
All you need to connect (with Windows authentication) now is the following code:
library(dplyr)
conn <- RSQLServer::src_sqlserver("servername", database = "dbname")
Limitations
Currently dplyr cannot deal with schema names in code like this:
conn %>% tbl("schema.table")
In this case you have to use
conn %>% tbl(sql("SELECT * FROM schema.table"))
But this issue is getting addressed in rstats-db/DBI/issues/24 (dplyr uses DBI).
What now?

Now that you have a connection to your SQL Server it's time to query all the data.
Check out the RStudio: Work with big data webinar or the dplyr databases vignette.
Have fun! 🙂


A nice and interesting post! You can also use an ODBC connection to connect with an SQL Server. For example, if you’re using a trusted connection to log in the SQL Server on your local or some remote machine, you could initialize the connection from R as follows:
library(RODBC)
conn % sqlQuery(‘select * from information_schema.tables’)
I’ve not previously familiarized myself with the RSQLServer package, but does it offer some benefits over a simple ODBC connection?
Hi JTT,
thanks for the hint, I haven’t tried it yet.
The advantage I see (but maybe I’m wrong), I can use RSQLServer with the package dplyr which has very convenient syntax for transforming data and querying tables (e.g. I don’t need to write a select statement).
Best regards,
Verena
Please note that the code in my original comment seems to have changed a bit, and will not work in that format. The idea anyhow was to be able to query SQL Server using dplyr as you showed on your article’s last code line, but that certainly needs an SQL query, and does not work with the out-of-box dplyr-functions such as select.
Did you try using RODBC library?
Hi, no I didn’t try that yet, thanks for the hint! I like about my approach that it works with dplyr which is really nice for transforming data.
Best,
Verena
I use RSQLServer to query SQL tables too! It definitely is a bit of a headscratcher to get everything set up and working correctly but I love using dplyr syntax to do the queries. I work on OSX so it’s different which parts to set up are hard; maybe I should put together a blog post on this…
Hi Julia, thanks for your comment! I use Windows at work and OS X at home. Luckily I could set it up on OS X without any issues at all. It probably won’t work with Windows user authentication but with username and password, right? I think a blog post about this would be great!